Welcome
Welcome to the Data Analysis and Coordination Center (DACC) for the Human Microbiome Project (HMP). The HMP was supported by the National Institutes of Health (NIH) Common Fund from 2007 through 2016. This area of the website focuses on the first of a two-phase effort, frequently referred to as HMP1, which ran from 2008 through 2013. From the Common Fund website:
The Human Microbiome Project has transitioned from Common Fund support. Common Fund programs are strategic investments that achieve a set of high-impact goals within a 5-10 year timeframe. At the conclusion of each program, deliverables transition to other sources of support or use by the broader scientific community. The HMP was supported by the Common Fund from 2007 to 2016. Non-HMP investment in microbiome research at the NIH has increased over forty-fold since the inception of the HMP and spans over 20 of the NIH Institutes and Centers.
Please note that since the HMP is no longer supported by the Common Fund, the program website is being maintained as an archive and will not be updated on a regular basis.
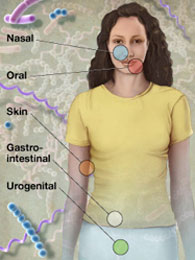
The overall mission of the HMP was to generate resources to facilitate characterization of the human microbiota to further our understanding of how the microbiome impacts human health and disease. The initial phase of the project, HMP1, established in 2008, characterized the microbial communities from 300 healthy individuals, across several different sites on the human body: nasal passages, oral cavity, skin, gastrointestinal tract, and urogenital tract. 16S rRNA sequencing was performed to characterize the complexity of microbial communities at each body site, and to begin to ask whether there is a core healthy microbiome. Metagenomic whole genome shotgun (wgs) sequencing provided insights into the functions and pathways present in the human microbiome. In total, over 32 terabytes of data were generated during this first phase of the project, all publicly available through the HMP Data Portal.
HMP1 was an interdisciplinary effort comprising four sequencing centers (the Broad Institute, Baylor College of Medicine, Washington University School of Medicine, and the J. Craig Venter Institute), a Data Analysis and Coordination Center (DACC), and several independent investigators involved in demonstration projects looking more closely at the associations between the microbiome and human health and disease, tool and technology development, and identification of ethical implications.
This site provides background information and resources associated with HMP1, including
- The Legacy HMP1 Data Browser, providing access to specific HMP1 datasets described in 2012 and 2017 landmark Nature papers.
- The HMP Data Portal, providing a repository of diverse human microbiome datasets from both HMP1 and the second phase of the project, iHMP, allowing researchers to query and retrieve metagenomic, metatranscriptomic, human genetic, microbial culture, and many other data types across both project.
If you are NEW TO EXPLORING HMP1 DATA or are interested in more details about project organization, we recommend you begin with the HMP1 Data Model.