Data Model
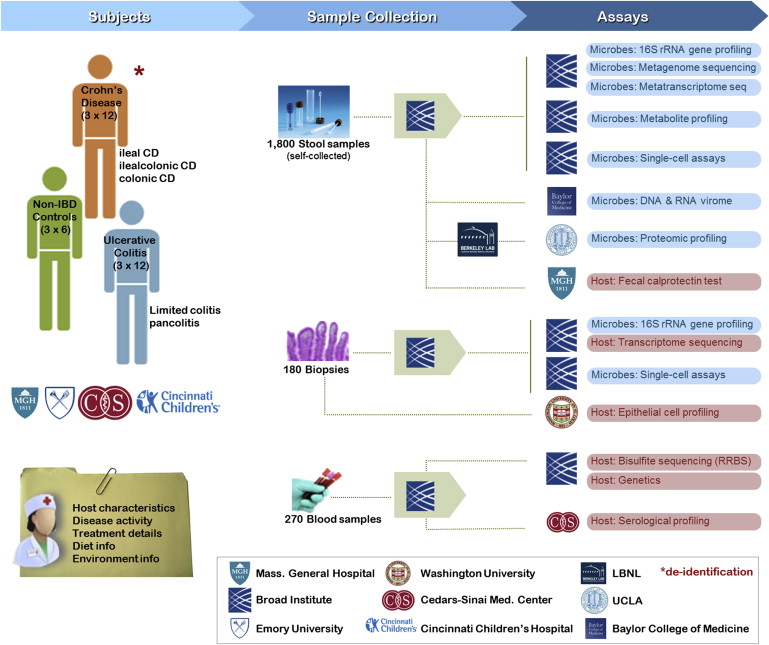
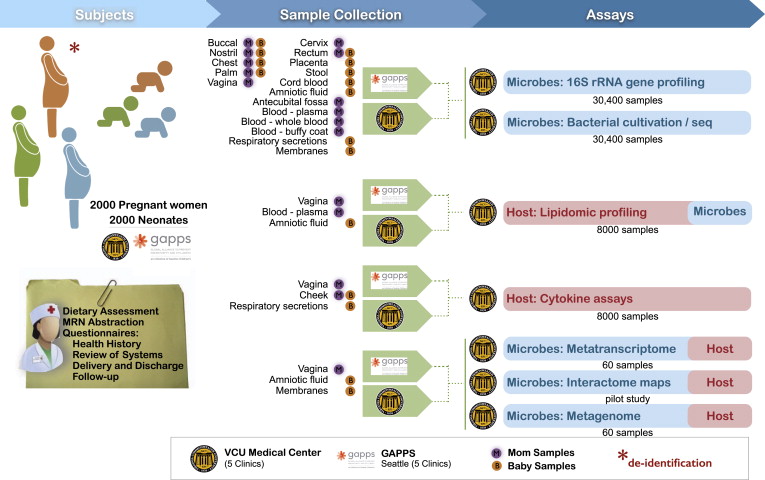
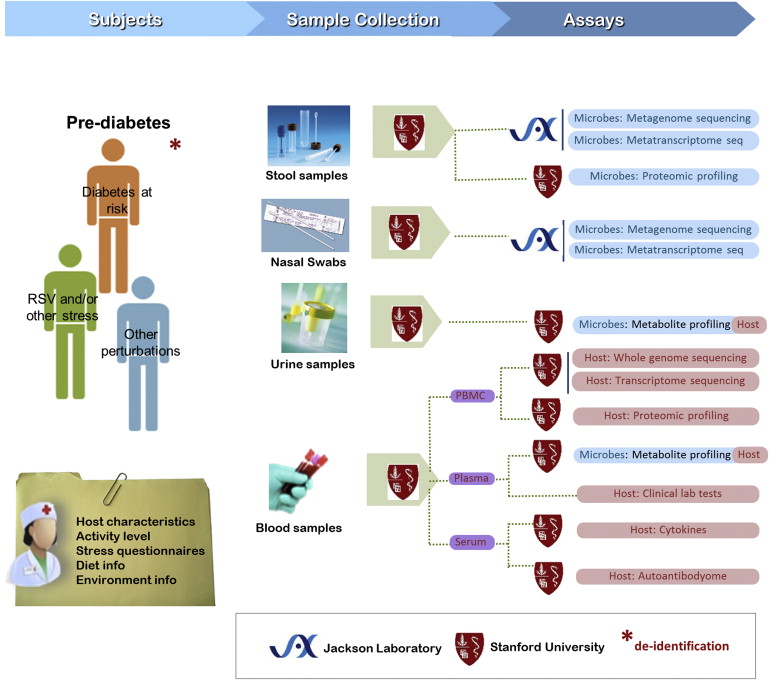
Much has been learned about the diversity and distribution of human-associated microbial communities, but we still know little about the biology of the microbiome, how it interacts with the host, and how the host responds to its resident microbiota. The iHMP is studying these interactions by creating integrated longitudinal datasets from both the microbiome and host from three different cohort studies of microbiome-associated conditions using multiple ‘omics technologies. Here we describe the three models of microbiome-associated human conditions, and the multi-omic data types to be collected, integrated, and distributed through public repositories as a community resource.
Public Repositories for primary multi-omic data
Source key: microbiome, host, global (host + microbiome)
| SRA | dbGap | GEO | EBI PRIDE | ATCC/BEI | Study DB | EBI IntAct | Metabolomics Workbench |
|---|---|---|---|---|---|---|---|
| 16S rRNA gene survey* | Subject genome sequences** | Whole transcriptome sequence | LC-MS/MS peptide profiles | Bacterial isolates | Cytokine profiles | Yeast two-hybrid binary protein complexes | Untargeted and targeted LC-MS metabolomic profiles |
| Whole metagenome shotgun sequences | Whole transcriptome sequences | Intestinal epithelial cell profiling response to bacterial metabolites | LC-MS/MS peptide profiles | Fecal calprotectin protein concentrations | Untargeted and targeted LC-MS lipidprofiles | ||
| Whole firome shotgun sequences | Human sequence from unfiltered total metagenomic sequences | Serology | |||||
| Bacterial single cell sequences | Subject phenotypes, clinical metadata, medical panels | ||||||
| Single cell bacterial transcript sequences | Intestinal epithelial cell profiling response to bacterial metabolites*** | ||||||
| Whole metatranscriptome shotgun sequences | |||||||
| Reduced representation bisulfite sequencing (RRBS) profiles |
Three models of microbiome-associated human conditional targeted by the iHMP
For more information, see the iHMP Marker Paper